-
The genusAeromonasis widely distributed in aquatic environments and is known to cause various diseases in humans and animals (1). These diseases include gastroenteritis, wound infections, bacteremia/sepsis, and immune dysfunction (2-3). Recently, certain motileAeromonasspecies have emerged as food- and waterborne pathogens, raising concern (4-5). Previous studies have reported that a majority (96.5%) of clinicalAeromonasisolates are identified asA. caviae(29.9%),A. dhakensis(26.3%),A. veronii(24.8%), andA. hydrophila(15.5%) (4,6). Of these,A. hydrophila,A. caviae, andA. veroniiare considered clinically significant (7-8).Aeromonasspecies cause severe, short-duration diarrheal disease or chronic loose stools, particularly in children, the elderly, and immunocompromised individuals (9), as well as traveler’s diarrhea (9-10).
In this study,Aeromonasstrains were isolated from adult patients with diarrhea and aquatic environments in Beijing Municipality, China. Whole-genome sequencing (WGS) and antibiotic resistance profiling were performed on theseAeromonasisolates. The findings of this research will provide valuable insights into the investigation of adult diarrhea caused byAeromonasin Beijing, China.
-
This study enrolled outpatients with acute diarrhea from two clinics in one district of Beijing, China, between January 1, 2016, and December 31, 2022, following the guidelines of the local adult diarrhea surveillance project in Beijing. Acute diarrhea was defined as at least three passages of watery, loose, mucous, or bloody stools within 24 hours. Patient age, sex, occupation, and clinical symptoms were recorded. Fresh fecal samples (5 mg) were collected from each patient, stored in Cary-Blair medium at 4 °C, and transported to the laboratory within 24 hours for bacterial isolation and real-time PCR detection. Additionally, in August 2023, water samples (1 L) were collected from the mainstream of the Chaobai River and its tributaries in Beijing, which flow through the study area. All collection sites were located away from factories and farms. Samples were stored in sterile containers and transported to the laboratory forAeromonasisolation within 2 hours.
Stool samples from patients were cultured to isolate various pathogens, includingAeromonasspp.,Salmonella,Shigella, diarrhealEscherichia coli,Vibrio parahaemolyticus,Vibrio cholerae,Campylobacter jejuni,Campylobacter coli, andYersinia enterocolitis(11). Real-time PCR was performed for the detection of norovirus, rotavirus, human sapovirus, adenovirus, and astrovirus using corresponding commercial kits (Beijing Applied Biological Technologies Co., Ltd., China). River water samples were cultured to isolate onlyAeromonasspp. For bacterial isolation, 200 mg of each patient stool sample was inoculated in alkaline peptone water (APW) and enriched at 37 °C for 24 h. Alternatively, 20 mL of 10× APW was added to river water samples and enriched at 37 °C for 24 h. After enrichment, a loopful of culture was streaked on MacConkey (MAC) agar and Rimler-Shotts (RS) agar and incubated at 37 °C for 24 h. Five presumptiveAeromonascolonies from the selective agar plate were selected and inoculated in tryptic soy agar (TSA) and incubated at 37 °C for 24 h. Each pure colony culture was initially identified using MALDI-TOF MS (Bruker).
Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA) following the manufacturer’s instructions. DNA samples (total amount >20 µg) were sent to the Beijing Genomics Institute for next-generation sequencing. The DNA concentration reached 50 ng/µL, and the OD260/280 ratio was 1.8–2.0 for individual samples. Low-quality reads were discarded, and clean data were assembled into genomic contigs using SOAPdenovo version 2.04. After removing contigs shorter than 500 bp, QUAST version 5.0.1 was used to assess the quality of the assembled genomes (Gurevich et al., 2013). Next, Prokka version 1.12 (Seemann, 2014), Prodigal version 2.6.3 (Hyatt et al., 2010), and RAST (https://rast.nmpdr.org/) were used to annotate the genomes. The genome sequences ofAeromonasisolates and seven strain types (A. hydrophilaATCC 7966T [accession No.: GCA_000014805],A. caviaeNCTC 12244T [GCA_900476005],A. dhakensisTN14 [GCA_905132925],A. veroniiFDAARGOS_632 [GCA_008693705],A. mediaCECT 4232T [GCA_000819985],A. sanarelliiLMG 24682T [GCA_000820085],A. schubertiiATCC 43700T [GCA_001481395]) were included in this study. ANI analysis was used to determine the evolutionary distance among the bacterial isolates at the genomic level. ANI values >95% were considered indicative of the same species. Prokka version 1.12 and the Roary genome pipeline with an identity cutoff of 95% were used to perform gene annotation and pan-genome analysis, respectively. Snippy was used to identify core genome single nucleotide polymorphisms (cgSNPs), with the genome sequence ofA. hydrophilaATCC 7966T used as the reference. The phylogenetic evolutionary tree was constructed based on decombined cgSNPs by IQ-TREE using maximum likelihood estimation with 1,000 bootstraps. The genome sequences of theAeromonasstrains sequenced in this study have been deposited at GenBank/DDBJ/ENA under the BioProject ID No. PRJNA1097280.
Antimicrobial susceptibility testing (AST) was then performed forAeromonasusing a microbroth dilution method and a panel for aerobic, gram-negative bacilli (Shanghai Fosun Long March Medical Science Co., Ltd., China). Antibiotic susceptibility (S), intermediate resistance (I), and resistance (R) were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines M45A2E (CLSI, 2019).Escherichia coliATCC 25922 was used as a control. The following antimicrobials were tested: ampicillin (AMP), ampicillin-sulbactam (AMS), tetracycline (TET), meropenem (MEM), polymyxin E (CT), ceftazidime/avibactam (CZA), cefotaxime (CTX), ceftazidime-avibactam (CAZ), ciprofloxacin (CIP), chloramphenicol (CHL), nalidixic acid (NAL), streptomycin (STR), sulfamethoxazole tablets (SXT), and amikacin (AMK).Aeromonasstrains were defined as multidrug-resistant (MDR) if they were resistant to three or more antimicrobial classes.
Statistical analyses were performed using Stata software (version 12.0). The chi-squared test was used to compare isolation rates between groups, with statistical significance set atP<0.05.
-
A total of 2,011 stool samples were collected from 2,011 patients with diarrhea between January 1, 2016, and December 31, 2022, excluding 2020, due to the COVID-19 pandemic. Of these, 13Aeromonasisolates (S1–S13), identified using MALDI-TOF MS, were derived from 13 patients (P1–P13), resulting in a positiveAeromonasrate of 0.65% (13/2,011). The annual positive rates were 0.27% (1/365) in 2016, 0% (0/371) in 2017, 0.53% (2/374) in 2018, 0.56% (2/358) in 2019, 0.84% (3/359) in 2021, and 2.72% (5/184) in 2022. These isolates were identified by their ANI values asA. veronii(n=5),A. caviae(n=5), andA. dhakensis(n=3;Table 1andFigure 1). Four of the 13Aeromonas-positive patients were co-infected with other intestinal pathogens: patient P1 withV. parahaemolyticus(tdh+/trh-); P2 withSalmonellaand EAEC; P3 withV. parahaemolyticus(tdh+/trh-); and P8 withV. parahaemolyticus(tdh+/trh-) and EPEC (Table 1).
Patient number Time of onset Sex Age Clinical symptoms Suspected contaminated
foodNumber of people dining together/
co- diners who experienced diarrheaBacterial culture results Number ofAeromonasisolate ANI analysis results ofAeromonas ANI value (%) Other pathogens detected P1 July 29, 2016 Male 26 Diarrhea (watery stools 10 times per day) + abdominal pain + fever
(38.1 ℃)Barbecue 1/- S1 A.caviae 97.936 Vibrio parahaemolyticus* P2 July 16, 2018 Female 30 Diarrhea (watery stools 10 times per day) + abdominal pain + nausea Cold meat 2/no S2 A.dhakensis 97.2863 Salmonella†+ EAEC P3 August 10, 2018 Male 44 Diarrhea (watery stools 12 times per day) + abdominal pain + nausea + vomiting Barbecue 1/- S3 A.dhakensis 97.179 V. parahaemolyticus* P4 June 25, 2019 Female 17 Diarrhea (loose stools 10 times per day) + abdominal pain + nausea + dehydration Fried food 6/yes S4 A.caviae 97.9062 / P5 July 16, 2019 Female 30 Diarrhea (loose stools 10 times per day) + abdominal pain + nausea Cold meat 3/no S5 A.dhakensis 97.0422 / P6 April 23, 2021 Male 81 Diarrhea (watery stools 10 times per day) + abdominal pain + nausea + weak + dehydration + vomiting Cold meat 1/- S6 A.caviae 97.9036 / P7 June 3, 2021 Female 20 Diarrhea (loose stools 6 times per day) + abdominal pain + nausea + weak + fever
(39.4 ℃)Salad 1/- S7 A.veronii 96.4188 / P8 July 26, 2021 Male 38 Diarrhea (watery stools 8 times per day) + abdominal pain + nausea Seafood 1/- S8 A.veronii 96.5328 V. parahaemolyticus*+EPEC P9 June 13, 2022 Male 40 Diarrhea (watery stools 10 times per day) + abdominal pain + nausea Cold drink 1/- S9 A.caviae 97.8955 / P10 August 4, 2022 Male 35 Diarrhea (loose stools 5 times per day) + abdominal pain Cold meat 3/yes S10 A.caviae 97.9052 / P11 August 13, 2022 Female 16 Diarrhea (watery stools 7 times per day) + abdominal pain Take-away fast food§ 5/yes¶ S11 A.veronii 96.5436 / P12 August 13, 2022 Female 17 Diarrhea (watery stools 5 times per day) + abdominal pain Take-away fast food§ 5/yes** S12 A.veronii 97.4809 / P13 August 27, 2022 Male 41 Diarrhea (watery stools 6 times per day) + abdominal pain + nausea + vomiting Cold meat 1/- S13 A.veronii 96.4993 / Abbreviation: ANI=average nucleotide identity; EAEC=enteroaggregativeEscherichia coli; EPEC=enteropathogenicEscherichia coli
*The virulence gene characteristics ofVibrio parahaemolyticusaretdh+/trh-;
†serotype isSalmonellaTyphimurium;
§specific foods include duck meat, chicken meat, broccoli, rice, boiled corn, tomato and egg soup, and apples;
¶P12 is one of the five co-diners of P11;
**P11 is one of the five co-diners of P12.Table 1.Information and clinical symptoms of patients with positive isolation ofAeromonas.
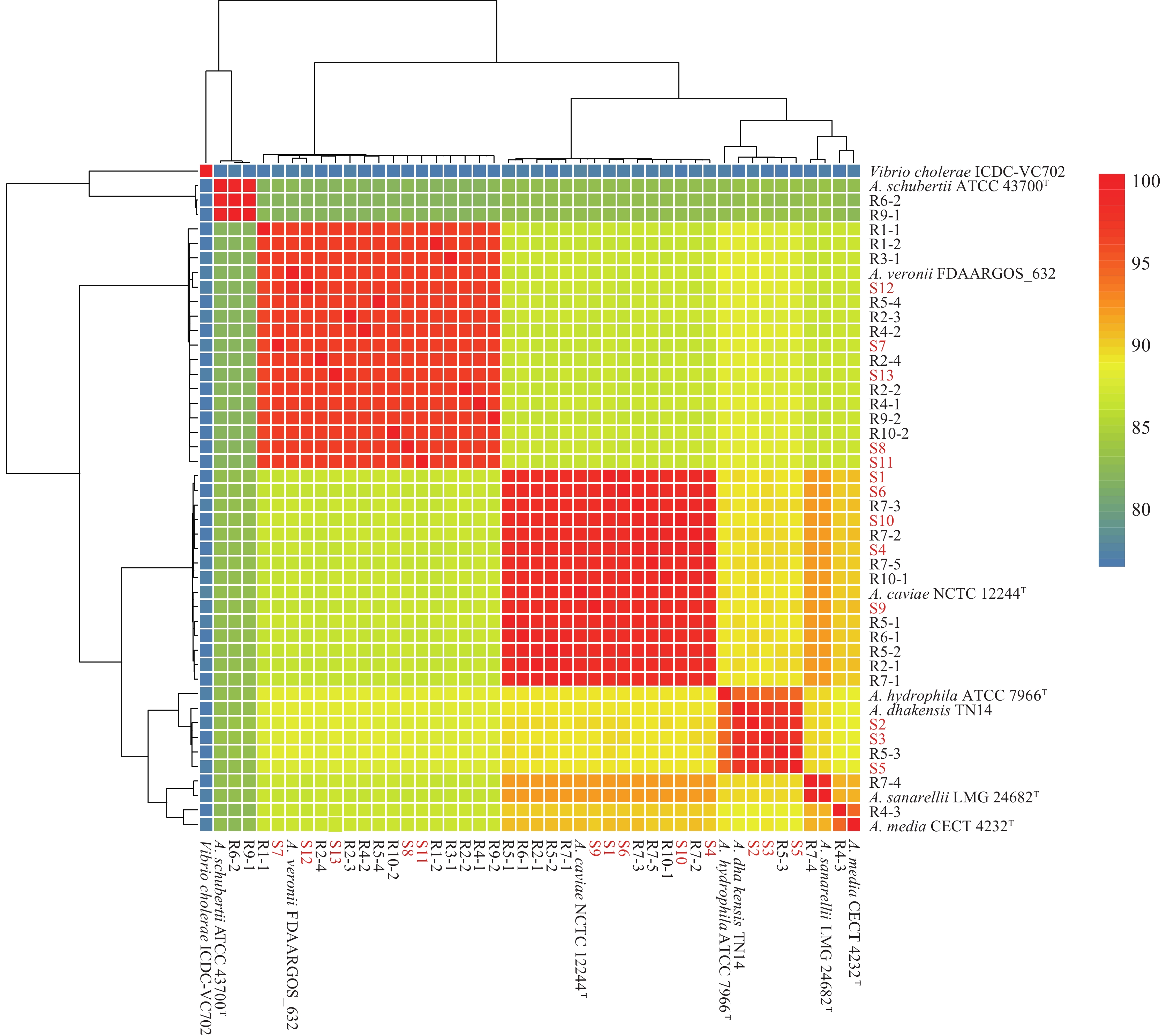 Figure 1.
Figure 1.Average nucleotide identity analysis ofAeromonasisolates.
Note: The red font represents strains from patients with diarrhea. T=type strain.Among the 13Aeromonas-positive patients, 7 were male and 6 were female, aged 16 to 81 years. Disease onset occurred in April (n=1), June (n=3), July (n=4), and August (n=5;Table 1). In the epidemiological investigation, all 13 patients reported consuming suspected contaminated food products, including cold meat, barbecue, fried food, salad, seafood, cold drinks, and takeaway fast food (Table 1). The most common clinical symptoms were diarrhea (100%, 13/13), abdominal pain (92.30%, 12/13), nausea (61.54%, 8/13), vomiting (23.08%, 3/13), weakness (15.38%, 2/13), dehydration (15.38%, 2/13), and fever (15.38%, 2/13). Moreover, 69.23% (9/13) and 30.77% (4/13) of patients presented with watery and loose stools, respectively (Table 1). Patients P11 and P12 were 2 students from the same school, with the same disease onset time (August 13, 2022). On this day, 5 individuals (including P11 and P12) at this school experienced diarrhea after consuming takeaway fast food delivered by the same catering company. All 5 patients presented with diarrhea (watery stools) and abdominal pain (Table 1).
Ten aquatic environmental samples (named A1–A10) were collected in this study, from which 25Aeromonasisolates were identified using MALDI-TOF MS. One sample (A8) yielded no isolates; the other nine water samples yielded between 1 and 5Aeromonasisolates each. The isolates were further classified asA. veronii(n=11),A. caviae(n=9),A. schubertii(n=2),A. sanarellii(n=1),A. dhakensis(n=1), and a species closely related toA. media(n=1;Table 2andFigure 1).
River water sample Bacterial culture results Aeromonasisolate ANI analysis results ofAeromonas ANI value (%) A1 + R1-1 A. veronii 96.19 R1-2 A. veronii 96.48 A2 + R2-1 A. caviae 97.89 R2-2 A. veronii 96.38 R2-3 A. veronii 96.55 R2-4 A. veronii 96.48 A3 + R3-1 A. veronii 96.48 A4 + R4-1 A. veronii 96.49 R4-2 A. veronii 96.44 R4-3 Species closely related toA. media 94.03 A5 + R5-1 A. caviae 97.82 R5-2 A. caviae 97.94 R5-3 A. dhakensis 97.16 R5-4 A. veronii 96.50 A6 + R6-1 A. caviae 97.88 R6-2 A. schubertii 99.00 A7 + R7-1 A. caviae 97.91 R7-2 A. caviae 97.97 R7-3 A. caviae 97.87 R7-4 A. sanarellii 98.02 R7-5 A. caviae 97.84 A8 − / / / A9 + R9-1 A. schubertii 98.98 R9-2 A. veronii 96.53 A10 + R10-1 A. caviae 97.83 R10-2 A. veronii 96.40 +: represents the sample culture positive inAeromonas.
−: represents the sample culture negative inAeromonas.
Abbreviation: ANI=average nucleotide identity.Table 2.Distribution ofAeromonasfrom river water samples.
-
TheAeromonasisolates from patients exhibited high resistance to AMP (92.31%), AMS (84.62%), NAL (69.23%), STR (61.54%), and TET (38.46%). However, fewer than 30% of isolates resisted the other antibiotics. Eight isolates (61.54%, 8/13) were identified as MDR, with the most dominant resistance patterns being AMP+AMS+NAL+STR (15.38%) and AMP+AMS+NAL (15.38%;Table 3andFigure 2).
Antibiotic Diarrhea patient isolates (% P/T)* River sample isolates (% P/T) AMP 92.31 (12/13) 96.00 (24/25) AMS 84.62 (11/13) 96.00 (24/25) TET 38.46 (5/13) 24.00 (6/25) MEM 0 (0/13) 12.00 (3/25) CT 15.38 (2/13) 4.00 (1/25) ETP 23.08 (3/13) 24.00 (6/25) CZA 0 (0/13) 0 (0/25) CTX 0 (0/13) 12.00 (3/25) CAZ 0 (0/13) 0 (0/25) CIP 0 (0/13) 0 (0/25) CHL 7.69 (1/13) 8.00 (2/25) NAL 69.23 (9/13) 60.00 (15/25) SXT 23.08 (3/13) 8.00 (2/25) STR 61.54 (8/13) 32.00 (8/25) AMK 0 (0/13) 0 (0/25) Abbreviation: AMP=ampicillin; AMS=ampicillin-sulbactam; TET=tetracycline; MEM=meropenem; CT=polymyxin E; ETP=ertapenem; CZA=ceftazidime/avibactam; CTX=cefotaxime; CIP=ciprofloxacin; NAL=nalidixic acid; CAZ=ceftazidime-avibactam; CHL=chloramphenicol; STR=streptomycin; SXT=Sulfamethoxazole Tablets; AMK=amikacin.
* P=number of resistance isolates; T=total number of isolates.Table 3.Results of antibiotic sensitivity test forAeromonasspecies.
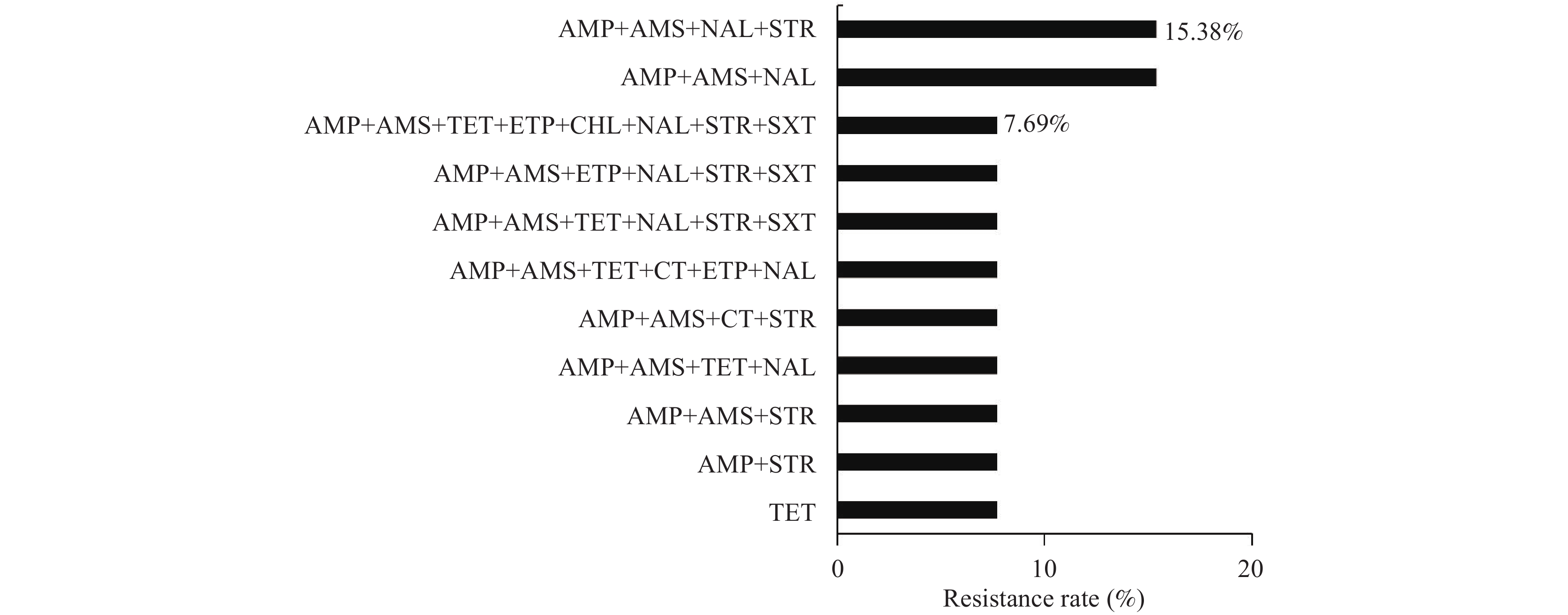 Figure 2.
Figure 2.Resistance spectrum of 13 strains ofAeromonasfrom patients to various antibiotic combinations.
Note: The X-axis represents the resistance rate of Aeromonas. The Y-axis represents a series of antibiotic combinations.TheAeromonasisolates from river water samples showed high resistance to AMP (96.00%), AMS (96.00%), NAL (60.00%), and STR (32.00%). Resistance to other antibiotics was observed in less than 30% of isolates. Eleven isolates (44.00%, 11/25) were MDR. The dominant resistance pattern was AMP+AMS (20.00%;Table 3andFigure 3).
-
A phylogenetic tree was constructed using cgSNP data from theAeromonasstrains with the reference sequenceA. hydrophilaATCC 7966T (Figure 4). The analysis revealed seven phylogenetic groups with a high bootstrap value. Seven species ofAeromonaswere located on their respective independent phylogenetic branches. Strains from patients were mainly distributed in the phylogenetic branches ofA. veronii,A. caviae, andA. dhakensis, while those from the river water samples were distributed in six different phylogenetic branches other than theA. hydrophilabranch. Of allAeromonasisolates obtained in this study, two isolates ofA. schubertii(R6-2 and R9-1) from the river water samples displayed genetic clustering; however, overall, there was no significant clustering observed between the isolates from patients and those from river water samples. TheAeromonasisolates from patients, and those from river water samples showed a significant distance between their phylogenetic branches even when they belonged to the same species and were derived from the same water sample. Moreover, the isolates from patients and the aquatic environment of the river water samples were intertwined in the phylogenetic tree.
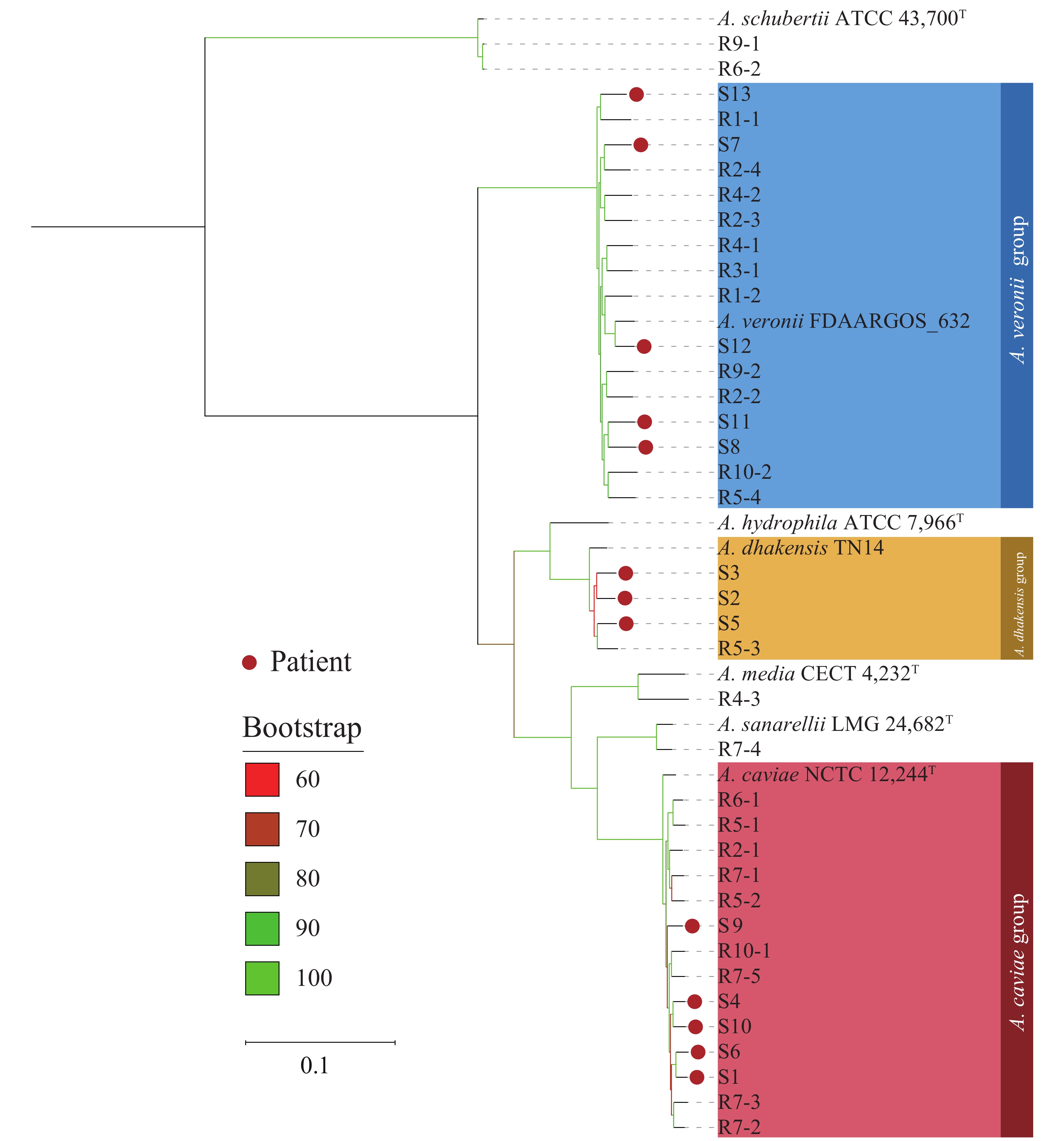 Figure 4.
Figure 4.Phylogenetic maximum likelihood tree based on core genome single nucleotide polymorphisms amongAeromonasstrains.
Note: Seven type strains or representative strains were also included. The three largest clusters ( A. veronii, A. caviae, A. dhakensis) are shown in blue, red, and yellow, respectively. The red dots on the top of the branches represent Aeromonasstrains isolated from patients with diarrhea. The various colors of branches (from red to green) represent bootstrap values ranging from 60% to 100%. T=type strain. -
Aeromonasspecies are recognized enteric pathogens, but their mechanisms of pathogenicity remain unclear. Previous studies from various locations have reported variable rates ofAeromonasdetection in patients with diarrhea. For instance, a study in Barcelona, Spain, reported a 2.09% (18/863) positive rate ofAeromonasin travelers with diarrhea, withA. veroniiandA. caviaeas the dominant species (12). Another study in Tainan City, Taiwan, China, also identifiedA. veroniiandA. caviaeas the dominant species in fish and clinical samples (13). Additionally, in Hong Kong Special Adiministrative Region, China, Chan et al. documented a 6.9% incidence ofAeromonasin adult patients with acute diarrhea (13). Isolation rates ofAeromonasfrom individuals with diarrheal illness in developed countries range from 0.8% to 7.4% (14). The most commonly isolated species implicated in gastroenteritis caused byAeromonasincludeA. veronii,A. hydrophila, andA. caviae, withA. trotaandA. jandaeioccurring less frequently (15). Similar to the aforementioned studies, our study isolatedAeromonasfrom 13 (0.65%) of the 2,011 patients with diarrhea, withA. veroniiandA. caviaebeing the most frequently isolated species.
In a previous study, gastroenteritis attributed toA. sobriawas characterized by acute abdominal pain, vomiting, diarrhea (watery stools), and fever (16), similar to the clinical symptoms of the patients enrolled in the present study. All 13Aeromonas-positive patients experienced abdominal pain and diarrhea, with diarrhea occurring 5–12 times per day and characterized by watery or loose stools. This was similar to the symptoms and stool characteristics caused byV. parahaemolyticus, a widely distributed pathogenic bacterium in aquatic products (17). Although the genusAeromonaswas initially considered to belong to the family Vibrionaceae, successive phylogenetic analyses have indicated thatAeromonasis not closely related to the genusVibrio, resulting in the reassignment ofAeromonasfrom the family Vibrionaceae to a new family (18). Notably, in the present study, all 13Aeromonas-positive patients reported consuming suspected contaminated food, with cold meat products being the most commonly reported (38.46% of patients). Cold meat products are associated with a high risk of foodborne diseases due to cross-contamination between raw and cooked products, which has also been observed in outbreaks caused byV. parahaemolyticusin China (19–22). In the present study,V. parahaemolyticuswas co-detected in 23.08% ofAeromonas-positive patients. The similarities betweenAeromonasandV. parahaemolyticusregarding their environmental distribution, association with suspected contaminated food, and clinical symptoms, as well as the co-infection ofAeromonasandV. parahaemolyticus, warrant further investigation. It is important to note that the subjective nature of patient-reported information about suspicious contaminated food is a limitation of the present study.
Aeromonasspp. are transmitted through food and water, with the ability to survive and, rapidly reproduce at low temperatures, and resist chlorine-containing disinfectants (23). However, to date, large-scale foodborne or waterborne outbreaks caused byAeromonasspp. have not been reported (23). In the present study, 2 patients (P11 and P12) who dined together and consumed takeout food from the same catering company — along with 3 other individuals — experienced diarrhea symptoms on the same day, suggesting a potential outbreak. However, genomic analysis did not confirm a common clone or establish a clear link between the outbreak andAeromonasspp. Successful molecular tracing of diarrhea outbreaks associated withAeromonasspp. has been limited (16,24-25). Nevertheless, outbreaks caused by multipleAeromonasspp. in patients have been reported (26), indicating a need for additional data on pathogenic characteristics in such outbreaks.
In this study,A. veroniiandA. caviaewere the dominant species among the 25Aeromonasisolates recovered from environmental water samples. Isolates from both patients and river water exhibited high levels of multidrug resistance, particularly to AMP and AMS. Although not statistically significant, the resistance rates to TET, CT, STR, and SXT — 4 antibiotics commonly used in clinical settings — were slightly higher among isolates from patients than among those from river water. This finding suggests that clinical treatment may contribute to antibiotic resistance inAeromonas.
The high bootstrap values of the main branches in the phylogenetic tree, based on cgSNP analysis ofAeromonasisolates from patients and river water samples, indicate a stable and reliable topology. Few isolates clustered significantly, and none were clonal (except for isolates R6-2 and R9-1, identified asA. schubertii). Patient-derived isolates of the sameAeromonasspecies (e.g., S7, S8, S11, S12, and S13) and isolates from the same river water sample belonging to the same species (e.g., R7-2, R7-3, and R7-4) were phylogenetically distant, suggesting they did not originate from a recent common ancestor. The interspersed distribution of patient-derived and river water-derived isolates in the phylogenetic tree underscores the importance of active surveillance for foodborne pathogens.
This study provides valuable insights into the prevalence and characteristics ofAeromonasin patients with diarrhea and aquatic environments. However, the study has limitations, including the subjective nature of patient-reported information and challenges establishing causal links between outbreaks andAeromonas, highlighting areas for further investigation.
HTML
Distribution ofAeromonasin Patients with Diarrhea
Antibiotic Susceptibility ofAeromonasIsolates
Phylogenetic Analysis ofAeromonas
| Citation: |




 Download:
Download:







